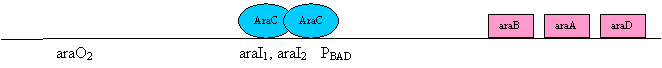|
|
Genetics Problem Spaces
|
|||||||||
|
Arabinose Operon Intro
Exercise Student Projects |
Arabinose
Operon Overview Arabinose is a 5 carbon sugar that can be used as an alternative carbon and energy source by the bacteria Escherichia coli. The enzymes necessary for the metabolism of this sugar are encoded by the araBAD operon. The operon encodes a single polycistronic mRNA with three open reading frames. Each open reading frame encodes one of the enzymes of the catabolic pathway for arabinose. Expression of the araBAD operon is highly regulated. Key to its regulation is the composition of sugars in the environment. For example, the operon shows a classic pattern of substrate level regulation. The operon is not expressed when arabinose is absent from the environment. It can only be expressed when arabinose is present in the environment. This is an adaptive mechanism that ensures the enzymes needed to catabolize arabinose are only produced when arabinose is present in the environment. The araBAD operon also exhibits the phenomena of catabolite repression. High levels of glucose in the environment prevent the expression of the araBAD operon even if arabinose is present. This is believed to be adaptive because bacteria can extract energy more efficiently from glucose than from arabinose. Therefore if glucose is present in the environment, the enzymes for metabolism of arabinose are not produced. The structure of the araBAD operon is diagramed
below. The three open reading frames,
araB, araA and
AraC is the regulatory protein that mediates substrate level regulation of transcription. It is a sequence specific binding protein that binds to the sequences at araI1, araI2 and araO2. Because AraC functions as a homodimer, it always binds two of these regulatory elements at the same time. AraC also has an arabinose binding pocket. The binding of arabinose to AraC alters is allosteric conformation, making the AraC protein more flexible. This increase flexibility is important to the regulation of the operon. AraC also has a domain that interacts with RNA polymerase, helping to recruit RNA polymerase to PBAD and thereby promoting transcription of araBAD. In the presence of arabinose, the AraC homodimer has a more flexible structure. This more flexible structure allows the AraC homodimer to simultaneously bind the nearby adjacent sites araI1 and araI2 (see diagram below). Binding of AraC to araI1/araI2 allows it to actively recruit RNA polymerase to PBAD and thereby promotes transcription.
In the absence of arabinose, the AraC homodimer has a ridged structure. This ridged structure interferes in binding to the closely adjacent sites araI1 and araI2. Instead the ridged AraC binds to araI1, and araO2. For a single homodimer of AraC to bind to these two distant DNA site requires DNA looping (see diagram below). The binding of AraC to these two sites prevents transcription of the araBAD operon in two different ways. First, since AraC is bound to araO2 instead of araI2, it is not able to promote recruitment of RNA polymerase. Second, the loop of DNA hinders the finding of the RNA polymerase to PBAD (as well as the binding of CAP to the cap binding site)
|