|
|
Genetics Problem Spaces
|
||
|
Arabinose Operon Intro
Exercise Student Projects |
Arabinose
Operon Introductory Exercise Introduction
An Investigation of
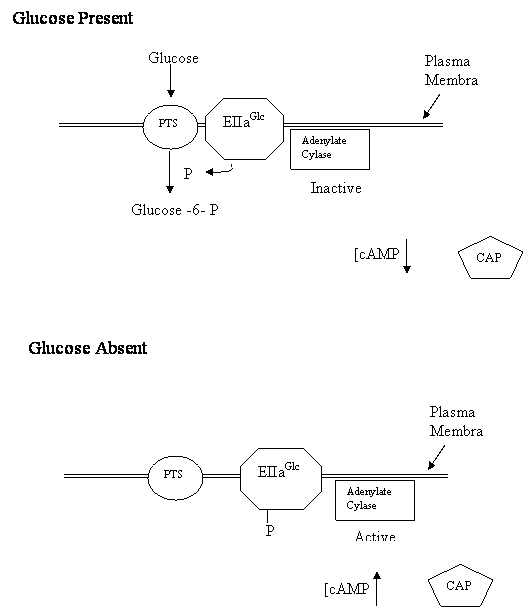
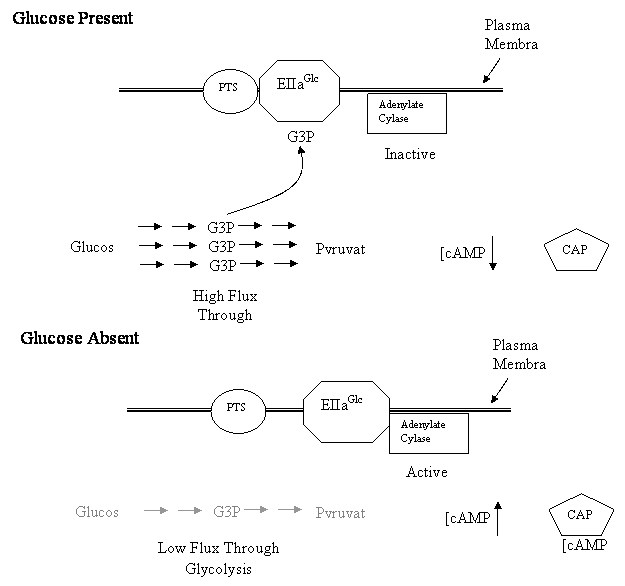
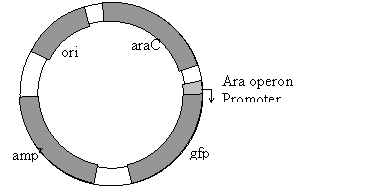
Two Models of Signal Transduction During Catabolite Repression Catabolite repression describes the phenomenon where certain carbon sources in bacterial growth media lead to a reduction of transcription of sensitive operons. In Escherichia coli, the best understood example of catabolite repression involves the lac operon. E. coli will not express the lac operon if glucose is present in the growth media. This is true whether or not lactose is present in the media. Other operons in E. coli are also repressed by glucose including the arabinose operon and the maltose operon. Although glucose is the most studied carbon source capable of catabolite repression in E. coli, it is not the only one capable of triggering repression of these operons. Sucrose, Fructose and other related carbon molecules can also trigger catabolite repression to varying levels. Klug and Cummings includes a discussion of the "classic cAMP" model for catabolite repression. In this model glucose acts by inhibiting the activity of adenylate cyclase. This leads to a drop in cAMP concentrations. At lower concentrations, the cAMP is not available to bind to the catabolite activator protein (CAP). CAP in the absence of cAMP loses its affinity for the Cap Binding Site and doesn't bind the lac operon. In the absence of CAP binding, the promoter is unable to recruit RNA polymerase to initiate transcription. Therefore, the lac operon is not transcribed in the presence of DNA. Alternatively, in the absence of glucose, the adenylate cyclase is not inhibited and actively produces cAMP. The cAMP binds to CAP causing it to shift to a conformation with affinity to the Cap Binding Site. CAP binds the Cap Binding Site and recruits the RNA polymerase to the promoter and initiates transcription of the lac operon. The "classic cAMP" model was proposed in the 1970's and its general acceptance by the scientific community is evidenced by it inclusion in all the genetics textbooks. However, in the last 5 years there has been a series of papers which challenge every aspect of the classic model. Several new models for catabolite repression have been proposed in these papers. Scientists in laboratories around the world are conducting new experiments to test each model. In this laboratory project, you will conduct an experiment to test an element of those models, the mechanism by which glucose inhibits adenylate cyclase. There are basically two models: the glucose transporter model and the glycolytic flux model. In the glucose transporter model, it is the movement of glucose through its membrane transport protein that triggers inhibition of adenylate cyclase (See Fig 1). In this model as glucose moves through the glucose transporter it is phosphorylated to form glucose - 6 - phosphate. The phosphate that is attached to glucose comes from a protein called EIIAglc. So that when glucose is moving through the transporter, the non-phosphorylated form of EIIAglc is more abundant. The non-phosphorylated form of EIIAglc does not interact with adenylate cyclase. In the absence of this interaction adenylate cyclase is inactive and the levels of cAMP decrease. In the glucose transporter model, when glucose is not moving through the glucose transporter, there is nothing to remove the phosphate from EIIAglc . The phosphorylated form of this protein accumulates in the cell. The phosphorylated form of EIIAglc interacts with adenylate cyclase activating it. The active adenylate cyclase makes cAMP and the levels of cAMP increase. In the glycolytic flux model, movement of glucose through the glucose transporter is not essential for catabolite repression. Instead, what is important is the flux of carbon through glycolysis (Fig 2.). Glycolysis is the metabolic pathway that catabolizes glucose to pyruvate. When glucose is being actively degraded by glycolysis, the concentration of phosphorylated intermediates of glycolysis such as glycerol-3-phosphate increase in the cell. In the glycolytic flux model, it is these phosphorylated intermediates that interact with EIIAglc and prevent EIIAglc from activating adenylate cyclase. Basically, the model says that abundant glucose leads to increase concentration of phosphorylated intermediates. These intermediates prevent EIIAglc from binding to adenylate cyclase and thereby prevent the activation of the enyzme. In the absence of activated adenylate cyclase, cAMP concentrations decrease. In the glycolytic flux model, when the cells are starved for glucose, the concentrations of glycolytic intermediates decrease. As their concentrations decrease, they are not available to interact with EIIAglc. This allows EIIAglc to activate the adenylate cyclase. Adenylate cyclase produces cAMP and the concentration of cAMP increase. For this laboratory, you will conduct an experiment to distinguish between these two models. The basic experimental strategy will be to test whether two carbon sources, galactose and glycerol, can trigger catabolite repression. What is important about these carbon sources is that they both increase glycolytic flux, but neither of them is transported through the glucose transporter. If the glucose transporter model were correct, then we would predict that neither of these compounds would effectively trigger catabolite repression. Since neither compound moves through the transporter, EIIAglc should remain in the phosphorylated state. And according to the glucose transporter model, this phosphorylated EIIAglc will activate adenylate cyclase and cAMP levels will rise. If the glycolytic flux model were correct, then we would predict that both of these compounds would trigger catabolite repression. The fact that they do not move through the glucose transporter is insignificant. What is important is that they both contribute to glycolysis (glycerol directly and galactose indirectly). Bacteria growing on these compounds should accumulate phosphorylated glycolytic intermediates. And according to the glycolytic flux model, these intermediates will prevent EIIAglc from activating adenylate cyclase and cAMP levels will decrease. In this experiment, we will not be measuring cAMP levels directly. Instead we will assay for it indirectly, by detecting catabolite repression. To assay catabolite repression we will use E. coli which contain engineered plasmids called pGLO (Fig 3). The pGLO plasmid contains a chimeric gene fusing the promoter elements from the arabinose operon to the jelly fish green fluorescent protein (gfp) open reading frame. The arabinose promoter contains a CAP Binding Site and shows the same type of catabolite repression as the lac operon promoter. Glucose prevents transcriptional initiation from this promoter even in the presence of arabinose. The gfp open reading frame is serving as a reporter gene. The expression of this protein is easy to detect because it fluoresces a bright green color under UV light. Bacteria lack the equivalent of this protein and typically don't fluoresce green under UV light. This means if the arabinose promoter of pGLO is active, the cells will fluoresce green. However, if the arabinose promoter is repressed the cells will not fluoresce. |
||
|
Experiment 1. Obtain overnight cultures of E. coli containing the pGLO plasmids from the teaching assistant. 2. Streak out the bacteria on LB media supplemented with the following carbon sources. LB + no supplements LB + 1% arabinose LB + 1% arabinose + 1% glucose LB + 1% arabinose + 1% glycerol LB + 1% arabinose + 1% galactose 3. Grow the bacteria overnight at 37°. 4. Store bacterial cultures at 4° until next laboratory period. 5. Visualize gfp expression under UV light. Short Report 1. Prepare a table reporting your results 2. Based on your results, what conclusions can you draw concerning whether the glucose transporter or glycolytic flux is involved in inhibiting adenylate cyclase. 3. Suppose that you had made a second set of plates with the same compounds as before, but that you had added 1% cAMP to the media to each. How would the results have changed? References Biorad (2000) Instruction Manual, Biotechnology Explorer Bacterial Transformation, the pGLO System Curriculum, Rev E. Available at <www.bio-rad.com/LifeScience/pdf/Bulletin_9563.pdf> (Retrieved on April 5, 2003) Brückner R, Titgemeyer F (2002) Carbon catabolite repression in bacteria: choice of the carbon source and autoregulatory limitation of sugar utilization. FEMS Microbiol Letters 209: 141-148. Eppler T, Postma P, Schültz A, Volker W, Boos W (2002) Glycerol-3 phosphate induced catabolite repression in Escherichia coli. J. Bacteriology 184: 3044-3052. Kimata K, Takahashi H, Inada , Postma P, Aiba H (1997) cAMP receptor protein-cAMP plays a crucial role in glucose-lactose diauxie by activating the major glucose transporter gene in Escherichia coli. PNAS 94: 12914-12919. Kimata K, Tanak Y, Inada T, Aiba H (2001) Expression of the glucose transporter gene, ptsG is regulated at the mRNA degredation step in response to glycolytic flux in Escherichia coli. EMBO J 20:3587-3595. Klug WS, Cummings MR (2003) Concepts of Genetics 7th ed. Prentice Hall, Upper Saddle River, New Jersey. pp. 449-459. Stülke J, Hillen W (1999) Carbon catabolite repression in bacteria. Curr Opin Microbiol 2:195-201. |
|||
|
Figure 1: Glucose transporter model Proteins Involved Glucose phosphotransferase system (PTS) EIIaGlc Adenylate Cyclase Catabolite Activator Protein (CAP)
|
|||
|
Figure 2: Glycolytic Flux Model Proteins Involved Glycolytic Enzymes EIIaGlc Adenylate Cyclase Catabolite Activator Protein (CAP)
|
|||
|
Figure 3: Map of pGLO plasmid
|